H12/UGS/3/5¶
This page describes a signal of selection found in the
Uganda An. gambiae populationusing the H12 (Garud et al. 20XX) statistic.The focus of this signal is on chromosome arm
3R between positions 36,460,000 and
37,520,000.
The evidence supporting this signal is
strong (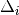 >= 100 on both flanks).
>= 100 on both flanks).
Signal location. Blue markers show the values of the selection statistic. The dashed black line shows the fitted peak model. The shaded red area shows the focus of the selection signal. The shaded blue area shows the genomic region in linkage with the selection event. Use the mouse wheel or the controls at the top right of the plot to zoom in, and hover over genes to see gene names and descriptions.
Genes¶
The following 71 genes overlap the focal region: AGAP009578 (adenosine monophosphate-protein transferase FICD homolog), AGAP009579 (dihydropyridine-sensitive l-type calcium channel), AGAP009580, AGAP009581 (collagen type I/II/III/V/XI/XXIV/XXVII alpha), AGAP009582, AGAP009583, AGAP0095841 (TRX1 - Thioredoxin), AGAP009585, AGAP009586, AGAP009587 (AP endonuclease 1), AGAP0095881 (alkylated DNA repair protein alkB homolog 4), AGAP009589, AGAP009590 (amiloride-sensitive sodium channel, other), AGAP0095911 (formyltetrahydrofolate dehydrogenase), AGAP009592, AGAP009593 (CP - Zinc carboxypeptidase A 1), AGAP009594 (Dynein assembly factor 1, axonemal homolog), AGAP009595 (sodium/potassium-transporting ATPase subunit beta), AGAP009596 (Sidestep protein), AGAP009597, AGAP028570, AGAP009598 (voltage-dependent calcium channel alpha-2/delta, invertebrate), AGAP0095994, AGAP009600 (ATP-dependent RNA helicase DOB1), AGAP009601 (RNA-binding protein 26), AGAP009602 (NADH dehydrogenase (ubiquinone) 1 beta subcomplex 6), AGAP009603 (SLC25A27 - mitochondrial uncoupling protein (solute carrier family 25)), AGAP009604, AGAP009605, AGAP009606, AGAP009607, AGAP009608, AGAP0096091 (homogentisate 1,2-dioxygenase), AGAP0096101 (glyoxylate/hydroxypyruvate reductase), AGAP0096111 (glyoxylate reductase/hydroxypyruvate reductase, gene 2), AGAP0096121 (glyoxylate/hydroxypyruvate reductase), AGAP009613 (mRpS7 - 28S ribosomal protein S7, mitochondrial), AGAP009614, AGAP009615 (U3 small nucleolar RNA-associated protein 13), AGAP009616, AGAP009617 (fatty-acid amide hydrolase 2), AGAP009618, AGAP009619 (S-adenosylmethionine decarboxylase proenzyme), AGAP009620 (reduction in cnn dots 4), AGAP009621 (alpha-N-acetylgalactosaminidase), AGAP009622 (6-phosphofructo-2-kinase / fructose-2,6-bisphosphatase), AGAP0096231 (GAPDH - glyceraldehyde 3-phosphate dehydrogenase), AGAP009624 (CTPsyn - CTP synthase), AGAP009625, AGAP009626 (Kv channel-interacting protein 4 isoform 2), AGAP009627, AGAP009628 (phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity), AGAP009629 (OBP5 - odorant-binding protein antennal 5), AGAP028743, AGAP028742, AGAP009631 (DNA topoisomerase 2-associated protein PAT1), AGAP009632 (ataxia telangiectasia mutated family protein), AGAP009633 (3-oxoacyl-(acyl-carrier protein) reductase fabG2), AGAP009634, AGAP009635, AGAP028571, AGAP009636, AGAP009637 (Bardet-Biedl syndrome 2 protein homolog), AGAP009638, AGAP009639 (nucleosome assembly protein 1-like 4), AGAP0096403 (Or1 - odorant receptor 1), AGAP009641 (kelch-like protein diablo), AGAP009642 (prohibitin 2), AGAP009643, AGAP009644 (DNA-directed RNA polymerases I and III subunit RPAC2), AGAP009645 (apoptosis inhibitor).
The following 3 genes are within 50 kbp of the focal region: AGAP009576 (collagen alpha 1), AGAP009577, AGAP009646 (Cad - homeobox protein caudal).
Key to insecticide resistance candidate gene types: 1 metabolic; 2 target-site; 3 behavioural; 4 cuticular.
Overlapping selection signals¶
The following selection signals have a focus which overlaps with the focus of this signal.
| Signal | Statistic | Population | Focus | Peak model  |
Max. percentile | Known locus |
|---|---|---|---|---|---|---|
| XPEHH/UGS.CMS/3/1 | XPEHH | Uganda An. gambiae | 3R:37,120,000-37,200,000 | 300 | 99.5% | nan |
| XPEHH/UGS.BFS/3/1 | XPEHH | Uganda An. gambiae | 3R:37,140,000-37,220,000 | 247 | 99.6% | nan |
Diagnostics¶
The information below provides some diagnostics from the Peak modelling algorithm.
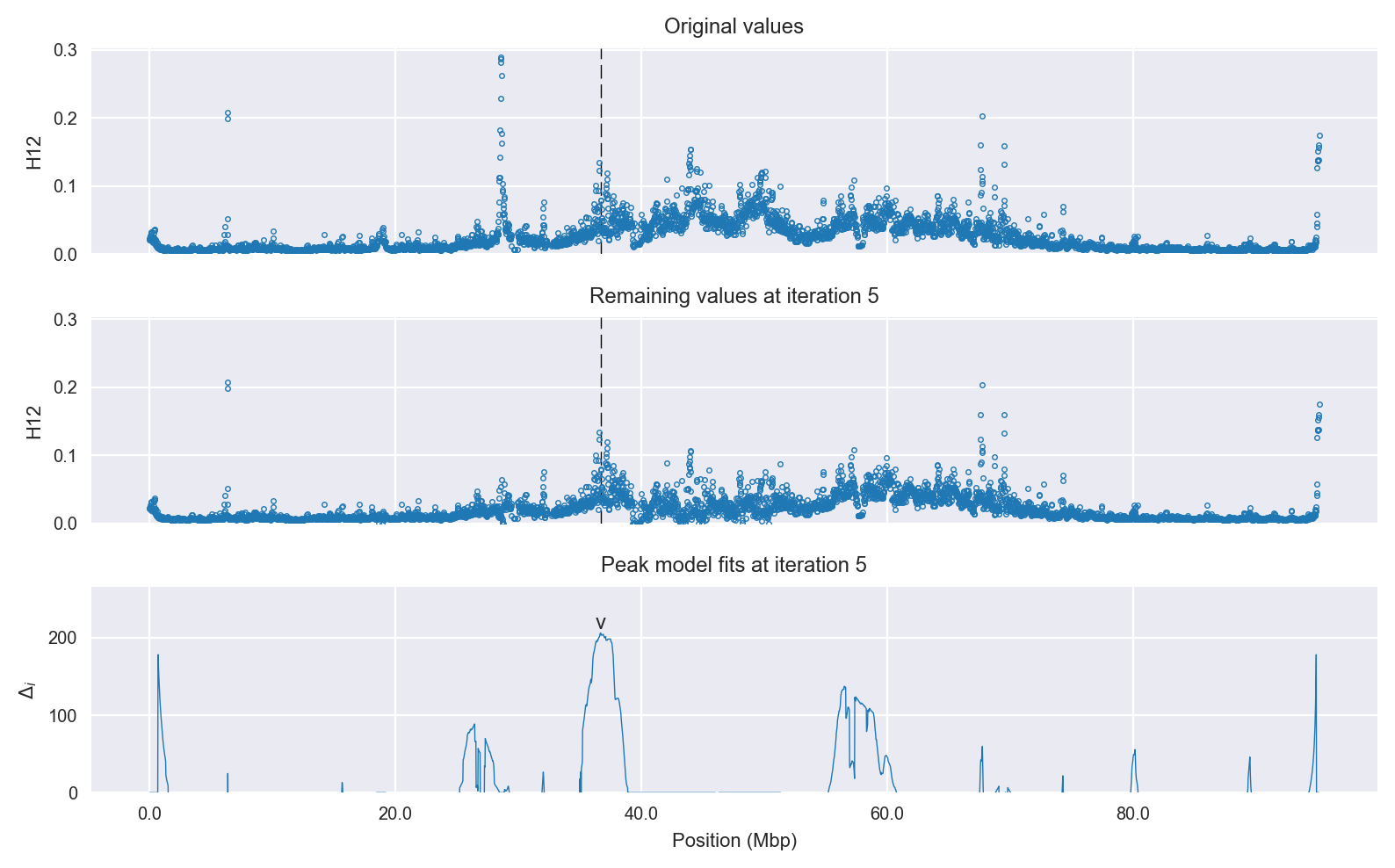
Selection signal in context. @@TODO
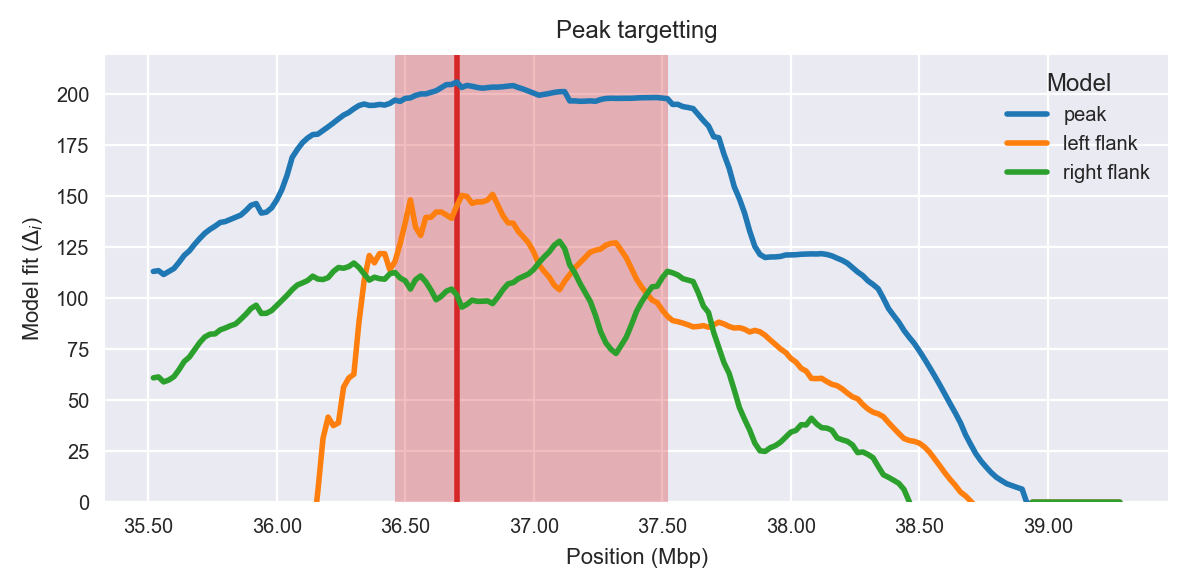
Peak targetting. @@TODO
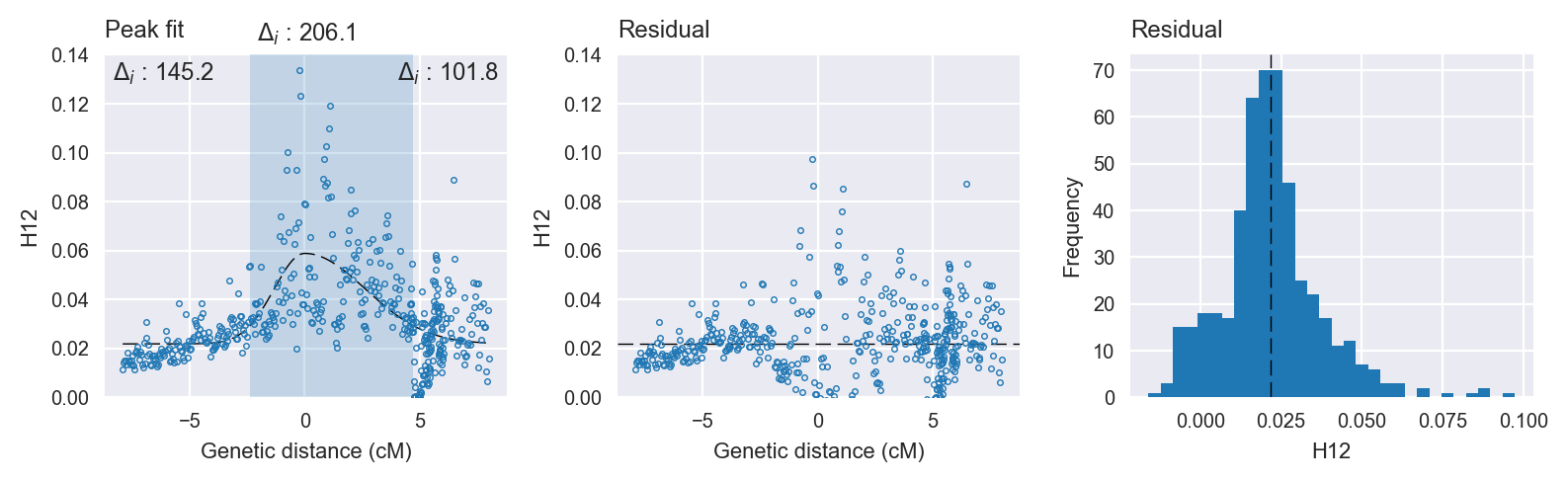
Peak fitting diagnostics. @@TODO
Model fit reports¶
Peak model:
[[Model]]
Model(skewed_gaussian)
[[Fit Statistics]]
# function evals = 48
# data points = 490
# variables = 4
chi-square = 0.117
reduced chi-square = 0.000
Akaike info crit = -4076.530
Bayesian info crit = -4059.752
[[Variables]]
center: 0 (fixed)
amplitude: 0.03699319 +/- 0.002177 (5.88%) (init= 0.5)
sigma: 1.87114700 +/- 0.148115 (7.92%) (init= 0.5)
skew: -0.49349950 +/- 0.074277 (15.05%) (init= 0)
baseline: 0.02182853 +/- 0.001297 (5.94%) (init= 0.03)
ceiling: 1 (fixed)
floor: 0 (fixed)
[[Correlations]] (unreported correlations are < 0.100)
C(sigma, baseline) = -0.647
C(amplitude, baseline) = -0.409
C(sigma, skew) = 0.351
C(amplitude, sigma) = -0.101
Null model:
[[Model]]
Model(constant)
[[Fit Statistics]]
# function evals = 9
# data points = 489
# variables = 1
chi-square = 0.178
reduced chi-square = 0.000
Akaike info crit = -3870.465
Bayesian info crit = -3866.272
[[Variables]]
c: 0.03191742 +/- 0.000863 (2.70%) (init= 0.03)